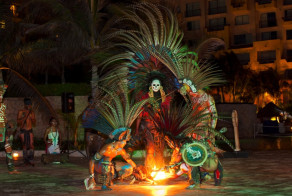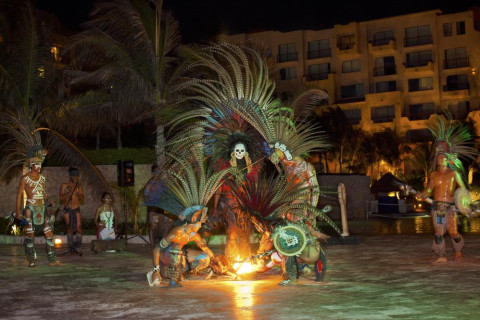- Home
- Past Conferences
- 3rd DNA Replication/Repair Structures and Cancer Conference
3rd DNA Replication/Repair Structures and Cancer Conference
11 Feb - 15 Feb 2018
Cancun, Mexico
Early Bird - Expired • Talk Submission - Expired • Poster Submission - Expired • Registration & Payment Deadline - Expired
Report
The 3rd DNA Replication/Repair Structures and Cancer Conference, which took place in Cancun, Mexico from 11-15 February 2018, centered on developing an actionable mechanistic knowledge of DNA replication and repair responses to stress for advanced cancer intervention strategies. Returning conference chairs, Sir Tom Blundell (FRS, FMedSci, Department of Biochemistry, University of Cambridge, UK) and Dr. John Tainer (Director SIBYLS Advanced Light Source, Professor and Director of Structural Biology, MD Anderson Cancer Center, Houston, Texas, USA) brought to Cancun many of the people driving advances in relevant structural biology and mechanisms, enabling uniquely productive and informative interactions for those working on DNA replication, damage responses and cancer. There was a total of 157 attendees who travelled worldwide to join us; Australia, USA, UK, The Netherlands, Japan, France, Saudi Arabia, China, South Korea, Germany, Italy, Poland, Canada, Sweden and Austria are just a few of the countries that were represented. Talks and poster sessions focused on cutting-edge structural and single-molecule approaches and novel mechanistic insights.
The remarkable stability of the human genome is lost in cancer cells due to the loss of efficient and accurate repair in the context of oncogene-induced replication stress and elevated transcription. DNA replication is furthermore emerging as a surprisingly fragile and complex process requiring fork protection and restart, R-loop resolution, and repair. Unresolved replication and repair intermediates signal apoptosis. The synthetic lethality and essentiality resulting from replication-repair stresses thus suggest repair inhibitors as tools to control pathway selection and damage outcomes and to design advanced therapeutics.
The 2018 meeting focused on structural and mechanistic insights into dynamic protein, DNA and RNA complexes acting in DNA replication and repair events relevant to cancer. The large diverse audience including many internationally distinguished speakers and the lively debates emphasised the importance of the theme. Comments from participants of all career stages emphasised their wish to see the series continue in this format and location.
“You and Tom did a really wonderful job putting together the conference with Laura and her colleagues. The science, discussions and location were all spot-on! I very much enjoyed participating in your conference and hope that I will have the opportunity to doing so again should you organise further events of this type.” Prof. Stephen Jackson (Wellcome Trust/Cancer Research UK Gurdon Institute)
The first day begun with a lively discussion of DNA Replication Machinery chaired by John Tainer and focused on structural features of high fidelity DNA replication and its enforcement. This was presented by an impressive line-up of international speakers. On this first day we had two breaks in the lecture schedule for sessions, expertly chaired by Susan Tsutakawa and Chris Brosey, of Meet the Poster Presenters, where all gave concise introductions and invitations to their poster. The evening concluded with an exciting plenary overview by Bruce Stillman from Cold Spring Harbor Laboratory on the structure, function and human genetics of the origin recognition complex and chromosome duplication.
The second day theme was replication fork stability, including mechanisms for maintaining genome stability at forks and for remodeling stalled replication forks. The following session focused further on sensing & signaling cellular genome instability. Mechanistic aspects were informed by amazing structural insights arising from state of the art techniques, for example of the structures of poly(ADP-ribose) polymerase enzymes. This session culminated in a brilliant plenary talk encompassing advances on cellular responses to DNA double-strand breaks by Steve Jackson of the Wellcome Trust/Cancer Research UK Gurdon Institute. The evening of the second day included sessions on defining & targeting the damage response. The final session, late in the evening but still with full attendance, was introduced by Tom Blundell, describing an opportunity to contribute to a themed volume of Progress in Biophysics and Molecular Biology, to be published by Elsevier.
These exciting sessions included breaks for discussions with refreshments & poster viewing, further sessions to ‘Meet the Poster Presenters’, and a short break for leisure in the afternoon. Some sought relaxing conversations on the beautiful beach 50 meters from the restaurants and lecture room! Discussions often further took place over dinner that could be taken at several lovely restaurants in the hotel.
The third day began with sessions on mutational signatures & mechanisms, focusing on mutagenesis in cancer, the concerted roles of genes in the protection of common fragile sites, the characterization of genomic rearrangements and DNA damage recognition. This session culminated with a fascinating plenary talk from Serena Nik-Zainal of the Wellcome Trust Sanger Institute. She detailed mechanistic insights and clinical applications of advances emerging from mutational signatures.
After a snorkeling expedition for some of the more energetic participants and informal afternoon discussions by others, the talks focused on DNA double-strand-break repair through Non-Homologous End Joining. This session was anchored by many strong talks and especially by the breakthrough new structure of the giant DNA-PK molecule, the largest single chain kinase in the human genome, which furthermore forms a point for assembly of many other components. New methods of investigating DNA end joining using proteomics-grade strategies for integrative structural biology and modelling were discussed, as well as exciting single-molecule analyses by Terence Strick measuring the time frame and energetics from the core and added components for this multicomponent DNA repair system.
The fourth day, Wednesday, started with talks and discussions of resection, repair and break signaling in homologous recombination of DNA double-strand breaks. One of the highlights was the way rad51 filament defines remodeling by human Rad51c-XRCC3. The discussion continued with a focus on DNA intermediates in repair and replication, and a discussion of mutagenesis, culminating with a very stimulating integrative plenary talk by Stephen West of the Francis Crick Institute on unresolved DNA intermediates as a source of DNA breaks and chromosome aberrations. The evening session continued with talks and discussions on coupling repair to replication and mitosis, with many stand out talks including those by Titia Sixma of the Netherlands Cancer Institute on structural biology of DNA mismatch repair and by Susan Lees-Miller of University of Calgary on non-repair functions of DNA-PKcs. All of this discussion included a look behind the scenes on publishing with the Nature research journals by Beth Moorefield that was followed by a gala dinner and poster awards (see below).
Despite the intensive schedule in talks and interactions, the final day continued with the same intensity of brilliant and exciting presentations on damage interactions with expression & chromatin. Talks included DNA damage sensor roles during base excision repair, structural analysis of protein DNA remodeler and structure and regulation of human ino80 complex by Dale Wigley. John Tainer from the MD Anderson Cancer Center completed the meeting with an exciting thought piece on the “rise of the molecular machines with cancer implications”.
The closing comments from the organisers expressed their happiness with the range of superb talks – probably the most intense five day meeting many have had for a long time and a great learning period. The meeting was attended by an enthusiastic and diverse audience, both with respect to stage of career, gender and nationality. We were proud to see that 20% of our attendees were students and this was due to the special rate offered by Fusion enabling many students to attend at cost price when accompanied by an academic.
The topic of DNA damage response and repair and its interface with replication remains an exciting area of research, still with many unanswered questions.
“Thank you, and all the organisers, so much for this wonderful opportunity to meet other researchers and make valuable contacts. I won't hesitate to say that this is one of the best conferences I've been to so far. Organisation is perfect and I will definitely want to attend in next edition…” Dr. Jacek Kabzinski (Medical University of Lodz)
Congratulations to all of those who received an award, details of which can be found below.
Organisers Short Talk Award
Noa Lamm-Shale (Children's Medical Research Institute)
Ravindra Amunugama (Harvard Medical School)
Poster Prize Awards
Zhe Yang (The Rockefeller University)
Taiana Maia de Oliveira (AstraZeneca)
Marcel Tuppi (Goethe-Universität Frankfurt)
Harshad Ghodke (University of Wollongong)
Benjamin Foster (IFE, Helmholtz Zentrum München)
Elizabeth O'Brien (California Institute of Technology)
FEBS Short Talk Award
Domi Baretic (MRC Laboratory of Molecular Biology)
Franz Klein (MFPL, University of Vienna)
We thank AstraZeneca for generously supporting this meeting and emphasising the importance of knowledge exchange between academia and industry, which is critical in our area and in this case can assist in the early development of new therapeutics for cancer. We also thank all our other wonderful sponsors; TESARO, Oncotarget, Repare Therapeutics, DECTRIS, artios, FEBS, Worldwide Cancer Research and the University of Pittsburgh. Their support was greatly appreciated and enabled us to fund many participants who otherwise may not have been able to attend. Special thanks to Elsevier for supporting the meeting and publishing the themed volume on selected talks.
The meeting was expertly organized, smoothly run, and supported by Fusion Conferences, Ltd. We thank Laura Trundle and her team for their helpful interactions with organisers and participants.
Synopsis
The remarkable stability of the human genome is lost in cancer cells due to the failure of efficient and accurate repair in the context of oncogene-induced replication stress and elevated transcription. DNA replication is furthermore emerging as a surprisingly fragile and complex process requiring fork protection and restart, template-switching, R-loop resolution, gap-filling, and repair. Unresolved replication and repair intermediates signal apoptosis. The synthetic lethality and essentiality resulting from replication-repair stresses thus suggest repair inhibitors as tools to control pathway selection and damage outcomes and to design advanced therapeutics.
The 3rd DNA Replication/Repair Structures & Cancer conference in February 2018 will focus on structural and mechanistic insights into dynamic protein, chromatin, DNA and RNA complexes acting in DNA replication and repair events relevant to cancer. Conference talks and discussions will center on developing an actionable mechanistic knowledge of DNA replication and repair stress responses suitable to guide advanced intervention strategies for cancer. The conference will emphasize an environment suitable for initiating productive collaborations. It will include vibrant informal and formal discussions together with informative talks and poster sessions.
To include late-breaking advances and innovating new investigators, one or more talks for every session will be selected from submitted abstracts. Meeting attendance is limited. Applicants seeking to present talks or posters are strongly encouraged to submit applications and abstracts early. All presenters are asked to focus on unpublished work, cutting-edge approaches and novel mechanistic insights that aid prediction of biological outcomes and plans to best apply basic research advances for cancer research and treatment.
What makes this meeting unique? Professor Ben Van Houten from the University of Pittsburgh says:
‘I can honestly say your meeting, by combining hard core structure-function studies with beautiful biology, hits an important area that many meetings miss; being part of that science is just terrific.’
Join the conference LinkedIn group to keep up to date with announcements and latest news concerning the conference.
Key Sessions
Sessions will include emerging results and breakthrough methods concerning:
- Repair and replication complexes, structures and mechanisms
- Mutational signatures, kataegis, DNA repair, and cancer
- Damage signaling, pathway selection & cross-talk
- Targeting synthetic lethality and essentiality for damage responses
- Chromatin and chromosome instability
- Epigenetics, DNA damage and repair
- PARP, PARG, and metabolic signaling
- Mitochondrial replication/repair and damage signaling
- RNA damage responses and actions in repair
- DNA repair, apoptosis and cancer
Confirmed Keynote Speakers
Steve Jackson (University of Cambridge)
'CELLULAR RESPONSES TO DNA DOUBLE-STRAND BREAKS'
Bruce Stillman (CSHL)
'THE ORIGIN RECOGNITION COMPLEX AND CHROMOSOME DUPLICATION: STRUCTURE, FUNCTION AND HUMAN GENETICS'
Steve West (The Francis Crick Institute)
'UNRESOLVED DNA INTERMEDIATES AS A SOURCE OF DNA BREAKS AND CHROMOSOME ABERRATIONS'
Serena Nik-Zainal (University of Cambridge)
'ADVANCES IN THE FIELD OF MUTATIONAL SIGNATURES: MECHANISTIC INSIGHTS AND CLINICAL APPLICATIONS'
Confirmed Invited Speakers
Lorena Beese (Duke University Medical Center)
Simon Boulton (The Francis Crick Institute)
'PITCH MODULATION AND EXTENSION DEFINES RAD51 FILAMENT REMODELLING BY HUMAN RAD51C-XRCC3 COMPLEX'
Chris Brosey (MD Anderson Cancer Center)
'BRIDGING DNA REPAIR AND NAD(H) ENERGETICS: TRACKING ARCHITECTURAL REGULATION OF APOPTOSIS-INDUCING FACTOR DURING PARTHANATOS'
Cynthia Burrows (University of Utah)
'DNA DAMAGE REGULATES GENE EXPRESSION VIA BASE EXCISION REPAIR'
Jean-Baptiste Charbonnier (University Parisâ€Saclay)
'ROLES OF Ku70/Ku80 IN NHEJ, FROM DSB RECOGNITION TO LIGATION'
Walter Chazin (Vanderbilt University)
'UNDERSTANDING HOW DNA PRIMASE WORKS REQUIRES KNOWLEDGE OF ITS ARCHITECTURAL DYNAMICS'
Karlene Cimprich (Stanford University School of Medicine)
'MECHANISMS FOR MAINTAINING GENOME STABILITY AT THE REPLICATION FORK'
David Cortez (Vanderbilt University)
'FUNCTIONS OF SINGLE-STRAND DNA BINDING PROTEINS IN MAINTAINING GENOME STABILITY'
Sheila David (University of California, Davis)
'BASE EXCISION REPAIR GLYCOSYLASES MUTYH AND NEIL1: THE GOOD, THE BAD AND THE DAMAGED'
Michelle Debatisse (Gustave Roussy Institute)
'COMMON FRAGILE SITE INSTABILITY: A RACE AGAINST TIME'
Sylvie Doublié (University of Vermont)
'STRUCTURAL ANALYSES OF DNA POLYMERASE THETA VARIANTS'
Structural analyses of DNA polymerase theta variants'
Brandt Eichman (Vanderbilt University)
'MECHANISMS OF REMODELING STALLED REPLICATION FORKS'
Dorothy Erie (University of North Carolina)
Ilya Finkelstein (University of Texas at Austin)
'ASSEMBLING THE HUMAN RESECTOSOME ONE MOLECULE AT A TIME'
Marco Foiani (University of Milan)
'AN INTEGRATED ATR, ATM AND mTOR-MECHANICAL NETWORK CONTROLLING NUCLEAR PLASTICITY AND CELL MIGRATIONB'
Mark Glover (University of Alberta)
'PNKP - A MEANS TO FIX THE ENDS'
Jane Grasby (Krebs Institute)
'COUPLING RECOGNITION AND REACTIVITY IN A STRUCTURE-SPECIFIC NUCLEASE'
Samir Hamdan (KAUST)
'SINGLE MOLECULE VIEW OF THE STRUCTURE SPECIFIC 5' NUCLEASES'
Reuben Harris (University of Minnesota)
'APOBEC MUTAGENESIS IN CANCER'
Karl-Peter Hopfner (Ludwig-Maximilians-University Munich)
'STRUCTURAL ANALYSIS OF PROTEIN DNA REMODELLER'
Ben Van Houten (University of Pittsburgh)
'DAMAGE SENSOR ROLE OF UV-DDB DURING BASE EXCISION REPAIR:STIMULATION OF APE1 AND OGG1'
Meindert Lamers (Leiden University Medical Center)
'STRUCTURAL FEATURES OF HIGH FIDELITY DNA REPLICATION'
Susan Lees-Miller (Arnie Charbonneau Cancer Institute)
'NON-REPAIR FUNCTIONS OF DNA-PKcs'
Karolin Luger (University of Colorado at Boulder)
'POLY (ADP-RIBOSE) POLYMERASE 1 IN DNA DAMAGE RECOGNITION'
Mauro Modesti (Cancer Research Center of Marseille)
'DYNAMICS AND MECHANICS OF DNA TETHERING BY CORE c-NHEJ FACTORS AND BY THE MRE11/RAD50/NBS1 COMPLEX'
Zachary Nagel (Harvard University)
'COMPREHENSIVE ANALYSIS OF DNA REPAIR CAPACITY IN NORMAL AND CANCEROUS CELLS'
Mark O'Connor (AstraZeneca)
'TARGETING THE REPLICATION STRESS RESPONSE IN CANCER'
John Pascal (Université de Montréal)
'STRUCTURAL BIOLOGY OF POLY(ADP-ribose) POLYMERASE ENZYMES'
Tanya Paull (University of Texas at Austin)
'THE Mre11-Rad50-Nbs1 COMPLEX AND REGULATION OF DNA REPAIR'
Phoebe Rice (University of Chicago)
'AN MCM-RELATED HELICASE AND OTHER REPLICATION-RELATED MACHINERY FOUND ON THE MOBILE ELEMENT BEHIND THE MRSA EPIDEMIC'
Andrej Sali (University of California, San Francisco)
'INTEGRATIVE STRUCTURE MODELING OF DNA-PKcs'
Orlando Scharer (Ulsan National Institute of Science and Technology)
'REPLICATIVE AND TRANSLESION SYNTHESIS DNA POLYMERASES IN ICL REPAIR'
Katharina Schlacher (MD Anderson Cancer Center)
'RAD51C IN DNA FORK PROTECTION AND STABILITY'
David Schriemer (University of Calgary)
'PROTEOMICS-GRADE STRATEGIES FOR INTEGRATIVE STRUCTURAL BIOLOGY'
Titia Sixma (NKI)
'STRUCTURAL BIOLOGY OF DNA MISMATCH REPAIR'
Maria Spies (University of Iowa)
'INHIBITORS OF THE RAD52-ssDNA INTERACTION'
Terence Strick (Institut Jacques Monod)
'SINGLE-MOLECULE ANALYSIS OF MULTICOMPONENT DNA REPAIR SYSTEMS'
Madalena Tarsounas (The CR-UK/MRC Oxford Institute for Radiation Oncology)
'WNT/b-CATENIN SIGNALING IN BRCA-DEFICIENT CELLS'
Susan Tsutakawa (Lawrence Berkeley National Laboratory)
'PHOSPHATE STEERING IS CRITICAL FOR CATALYSIS AND PREVENTION OF OFF-TARGET INCISIONS IN HUMAN FLAP ENDONUCLEASE'
Ashok Venkitaraman (University of Cambridge)
'INSIGHTS INTO CARCINOGENESIS FROM STRUCTURAL AND BIOLOGIC ANALYSES OF THE BRCA2 TUMOUR SUPPRESSOR'
Dale Wigley (Imperial College London)
'STRUCTURE AND REGULATION OF HUMAN INO80 COMPLEX'
Scott Williams (NIEHS)
'ENFORCEMENT OF DNA LIGATION FIDELITY'
Confirmed Speakers
Chairs
Plenary Speakers

Steve West
Principal Group Leader, Francis Crick Institute

Bruce Stillman
President and Professor, Cold Spring Harbor Laboratory

Stephen Jackson
Senior Group Leader, CRUK- Cambridge Institute

Serena Nik-Zainal
., University of Cambridge
Invited Speakers

Madalena Tarsounas
CR-UK Senior Group Leader, The CR-UK/MRC Oxford Institute for Radiation Oncology, University of Oxford

Marco Foiani
PI, IFOM ETS - the AIRC Institute of Molecular Oncology

Katharina Schlacher
Associate Professor, MD Anderson Cancer Center

Simon Boulton
Principal Group Leader, The Francis Crick Institute

Tanya Paull
Professor, The University of Texas at Austin

Andrej Sali
Professor, University of California, San Francisco

David Cortez
Professor, Vanderbilt University

Titia Sixma
Group leader, Netherlands Cancer Institute

Karolin Luger
Investigator, HHMI; Professor, CU Boulder, University of Colorado Boulder

Cynthia Burrows
Distinguished Professor, University of Utah

Walter Chazin
Professor and Senior Associate Dean; Founding Director, Center for Structural Biology, Vanderbilt University

Chris Brosey
Research Scientist, The University of Texas M.D. Anderson Cancer Center

Ilya Finkelstein
Associate Professor, UT-Austin

Susan Tsutakawa
Research Scientist, Lawrence Berkeley National Laboratory

Brandt Eichman
Professor, Vanderbilt University

Mark O'Connor
Chief Scientist, AstraZeneca

Mark Glover
Professor, University of Alberta

Sylvie Doublié
Professor, University of Vermont

Meindert Lamers
Associate Professor, Leiden University Medical Center

Mauro Modesti
Principal Investigator, Cancer Research Center of Marseille

David Schriemer
Professor, University of Calgary

Dorothy Erie
Professor, University of North Carolina

Maria Spies
Professor of Biochemistry and Molecular Biology, University of Iowa

Michelle Debatisse
Researcher, UMR8200 CNRS

Reuben Harris
Professor, University of Minnesota

Sheila David
Professor of Chemistry, University of California, Davis

Ben Van Houten
Richard M. Cyert Professor of Molecular Oncology, UPMC Hillman Cancer Center

Karlene Cimprich
Professor, Stanford University

Karl-Peter Hopfner
Professor, Gene Center

Jean-Baptiste Charbonnier
Team Leader, CEA-CNRS-Univ Paris Saclay, France

Orlando D. Schärer
Associate Director, Institute for Basic Science

Ashok Venkitaraman
Director, MRC Cancer Unit, University of Cambridge

Samir Hamdan
Assistant Professor of Bioscience, King Abdullah University of Science and Technology

John Pascal
Professor, Université de Montréal

Susan Lees-Miller
Professor, University of Calgary

Dale Wigley
Professor, Imperial College London

Zachary Nagel
Assistant Professor of Radiation Biology, Harvard U

Lorena Beese
Professor, Duke University Medical Center

Phoebe Rice
Professor, The University of Chicago

Scott Williams
Principal Investigator, NIH/NIEHS

Terence Strick
Research Director, Ecole normale supérieure/CNRS

Jane Grasby
Professor of Biological Chemistry, University of Sheffield
Programme
DISCUSSION IS VALUED: 5 MINUTES IS SAVED FOR DISCUSSION AFTER EACH TALK
SUNDAY 11TH FEBRUARY 2018 |
||
|
09:00 – 15:00 |
Group Trip: Tulum (signups required in advance) |
|
|
15:00 – 16:00 |
Registration & Welcome Reception |
|
|
16:00 – 16:10 |
Welcome and Opening Remarks by John Tainer & Tom Blundell |
|
|
SUNDAY PM |
DNA Replication Machinery |
|
|
16:10 – 16:30 |
Meindert Lamers |
STRUCTURAL FEATURES OF HIGH FIDELITY DNA REPLICATION |
|
16:35 – 16:45 |
Ravindra Amunugama * |
REPLICATION FORK REVERSAL DURING DNA INTERSTRAND CROSSLINK REPAIR REQUIRES CMG HELICASE UNLOADING |
|
16:50 – 17:00 |
Philipp Oberdoerffer * |
REPLICATION STRESS SHAPES A PROTECTIVE CHROMATIN ENVIRONMENT ACROSS FRAGILE GENOMIC REGIONS |
|
17:05 – 17:25 |
Samir Hamdan |
SINGLE MOLECULE VIEW OF THE STRUCTURE SPECIFIC 5' NUCLEASES |
|
17:30 – 17:50 |
Scott Williams |
|
|
17:55 – 18:20 |
Meet the Poster Presenters I |
|
|
18:20 – 18:50 |
Refreshments |
|
|
18:50 – 19:10 |
Meet the Poster Presenters II |
|
|
19:10 – 19:20 |
Tahir Tahirov * |
ELABORATED ACTION OF THE HUMAN PRIMOSOME |
|
19:25 – 19:45 |
Walter Chazin |
UNDERSTANDING HOW DNA PRIMASE WORKS REQUIRES KNOWLEDGE OF ITS ARCHITECTURAL DYNAMICS |
|
19:50 – 20:30 |
Plenary Speaker: |
THE ORIGIN RECOGNITION COMPLEX AND CHROMOSOME DUPLICATION: STRUCTURE, FUNCTION AND HUMAN GENETICS |
|
20:35 |
Dinner at Leisure |
|
MONDAY 12TH FEBRUARY 2018 |
||
|
07:00 – 08:15 |
Breakfast |
|
|
MONDAY AM |
Replication Fork Stability |
|
|
08:15 – 08:20 |
Session Introduction by Marco Foiani |
|
|
08:20 – 08:40 |
Karlene Cimprich |
MECHANISMS FOR MAINTAINING GENOME STABILITY AT THE REPLICATION FORK |
|
08:45 – 09:05 |
Brandt Eichman |
MECHANISMS OF REMODELING STALLED REPLICATION FORKS |
|
09:10 – 09:30 |
David Cortez |
FUNCTIONS OF SINGLE-STRAND DNA BINDING PROTEINS IN MAINTAINING GENOME STABILITY |
|
09:35 – 09:45 |
Martin Cohn * |
IDENTIFICATION OF UHRF2 (RNF107) AS A NOVEL ICL SENSOR PROTEIN REQUIRED FOR THE FANCONI ANEMIA PATHWAY |
|
09:50 – 10:10 |
Katharina Schlacher |
RAD51C IN DNA FORK PROTECTION AND STABILITY |
|
10:15 – 10:45 |
Refreshments & Poster Viewing |
|
|
MONDAY AM |
Sensing & Signaling Cellular Genome Instability |
|
|
10:45 – 10:50 |
Session Introduction by Katharina Schlacher |
|
|
10:50 – 11:10 |
Marco Foiani |
AN INTEGRATED ATR, ATM AND mTOR-MECHANICAL NETWORK CONTROLLING NUCLEAR PLASTICITY AND CELL MIGRATION |
|
11:15 – 11:25 |
Noa Lamm-Shalem * |
ATR AND mTOR REGULATE NUCLEUR ACTIN POLYMERIZATION IN RESPONSE TO REPLICATION STRESS TO ALTER NUCLEUR ARCHITECTURE AND MAINTAIN GENOME STABILITY |
|
11:30 – 11:50 |
John Pascal |
STRUCTURAL BIOLOGY OF POLY(ADP-ribose) POLYMERASE ENZYMES |
|
11:55 – 12:15 |
Chris Brosey |
BRIDGING DNA REPAIR AND NAD(H) ENERGETICS: TRACKING ARCHITECTURAL REGULATION OF APOPTOSIS-INDUCING FACTOR DURING PARTHANATOS |
|
12:20 – 13:00 *Partially sponsored by Worldwide Cancer Research* |
Plenary Speaker: |
CELLULAR RESPONSES TO DNA DOUBLE-STRAND BREAKS |
|
13:05 – 16:25 |
Lunch at Leisure & Free Time |
|
|
MONDAY PM |
Defining & Targeting The Damage Response |
|
|
16:25 – 16:30 |
Session Introduction by Susan Lees-Miller |
|
|
16:30 – 16:50 |
Mark O'Connor |
TARGETING THE REPLICATION STRESS RESPONSE IN CANCER |
|
16:55 – 17:15 |
Jane Grasby |
COUPLING RECOGNITION AND REACTIVITY IN A STRUCTURE-SPECIFIC NUCLEASE |
|
17:20 – 17:30 |
Mohiuddin * |
CTIP-BRCA1 COMPLEX AND MRE11 NUCLEASE PLAY A CRITICAL ROLE IN MAINTAINING REPLICATION FORKS IN THE PRESENCE OF CHAIN TERMINATING NUCLEOTIDE ANALOGS |
|
17:35 – 18:15 |
Meet the Poster Presenters III |
|
|
18:15 – 18:55 |
Poster Session & Refreshments (even poster numbers only) |
|
|
18:55 – 19:15 |
Madalena Tarsounas |
WNT/b-catenin signaling in BRCA-deficient cells |
|
Maria Spies |
INHIBITORS OF THE RAD52-ssDNA INTERACTION |
|
|
19:45 – 19:55 |
Mona Al-Mugotir * |
TARGETING RAD52 PROTEIN-PROTEIN INTERACTIONS IN CANCER THERAPY |
|
20:00 – 20:20 |
Zachary Nagel |
COMPREHENSIVE ANALYSIS OF DNA REPAIR CAPACITY IN NORMAL AND CANCEROUS CELLS |
|
20:25 – 20:40 |
Publication Opportunity: Progress in Biophysics and Molecular Biology (Published by Elsevier) Announcement by Tom Blundell |
|
|
20:40 |
Dinner at Leisure |
|
TUESDAY 13TH FEBRUARY 2018 |
||
|
07:00 – 08:15 |
Breakfast |
|
|
TUESDAY AM |
Mutational Signatures & Mechanisms |
|
|
08:15 – 08:20 |
Session Introduction by Bennett Van Houten |
|
|
08:20 – 08:40 |
Reuben Harris |
APOBEC MUTAGENESIS IN CANCER |
|
08:45 – 08:55 |
Agnel Sfeir * |
POLYMERASE THETA PROMOTES ERROR-PRONE REPAIR BY alt-NHEJ AT THE EXPENSE OF ERROR-FREE HR |
|
09:00 – 09:10 |
Xiaohua Wu * |
THE CONCERTED ROLES OF FANCM AND Rad52 IN THE PROTECTION OF COMMON FRAGILE SITES |
|
09:15 – 09:35 |
Ashok Venkitaraman |
INSIGHTS INTO CARCINOGENESIS FROM STRUCTURAL AND BIOLOGIC ANALYSES OF THE BRCA2 TUMOUR SUPPRESSOR |
|
09:40 – 10:10 |
Group Photo, Refreshments & Poster Viewing |
|
|
10:10 – 10:20 |
Anne Bothmer * |
CHARACTERIZATION OF GENOMIC REARRANGEMENTS IN RESPONSE TO CRISPR/Cas9-INDUCED DOUBLE-STRAND BREAKS |
|
10:25 – 10:45 |
Susan Tsutakawa |
PHOSPHATE STEERING IS CRITICAL FOR CATALYSIS AND PREVENTION OF OFF-TARGET INCISIONS IN HUMAN FLAP ENDONUCLEASE |
|
10:50 – 11:10 |
Karolin Luger |
POLY (ADP-RIBOSE) POLYMERASE 1 IN DNA DAMAGE RECOGNITION |
|
11:15 – 11:35 |
Michelle Debatisse |
COMMON FRAGILE SITE INSTABILITY: A RACE AGAINST TIME |
|
11:40 – 12:20
|
Plenary Speaker: |
ADVANCES IN THE FIELD OF MUTATIONAL SIGNATURES: MECHANISTIC INSIGHTS AND CLINICAL APPLICATIONS |
|
12:25 – 16:30 |
Lunch at Leisure & Free Time |
|
|
12:40 – 16:00 |
Group Trip: Snorkeling (signups required in advance) |
|
|
TUESDAY PM |
Non-Homologus End Joining |
|
|
16:30 – 16:35 |
Session Introduction by Simon Boulton |
|
|
16:35 – 16:55 |
Tom Blundell |
DNA-PKcs STRUCTURE FORMS A STAGE FOR THE ACTORS AND SUGGESTS AN ALLOSTERIC MECHANISM FOR MODULATING DNA DOUBLE-STRAND-BREAK REPAIR |
|
17:00 – 17:20 |
David Schriemer |
PROTEOMICS-GRADE STRATEGIES FOR INTEGRATIVE STRUCTURAL BIOLOGY |
|
17:25 – 17:35 |
Benjamin Stinson * |
NON-HOMOLOGOUS END JOINING OPERATES BY A “LIGASE-FIRST” MECHANISM
|
|
17:40 – 18:00 |
Terence Strick |
SINGLE-MOLECULE ANALYSIS OF MULTICOMPONENT DNA REPAIR SYSTEMS |
|
18:05 – 18:25 |
Andrej Sali |
INTEGRATIVE STRUCTURE MODELING OF DNA-PKcs |
|
18:30 – 19:10 |
Poster Session & Refreshments (odd poster numbers only) |
|
|
19:10 – 19:30 |
Jean-Baptiste Charbonnier |
ROLES OF Ku70/Ku80 IN NHEJ, FROM DSB RECOGNITION TO LIGATION |
|
19:35 – 19:55 |
Tanya Paull |
INTERSECTIONS BETWEEN MRN AND THE NON-HOMOLOGOUS END JOINING MACHINERY |
|
20:00 – 20:20 |
Mark Glover |
PNKP - A MEANS TO FIX THE ENDS |
|
20:25 |
Dinner at Leisure |
|
THURSDAY 15TH FEBRUARY 2018 |
||
|
07:00 – 08:15 |
Breakfast |
|
|
THURSDAY AM |
Damage Interactions With E xpression & Chromatin |
|
|
08:15 – 08:20 |
Session Introduction by Tom Blundell |
|
|
08:20 – 08:40 |
Bennett Van Houten |
DAMAGE SENSOR ROLE OF UV-DDB DURING BASE EXCISION REPAIR:STIMULATION OF APE1 AND OGG1 |
|
08.45 – 09:05 |
Cynthia Burrows |
DNA DAMAGE REGULATES GENE EXPRESSION VIA BASE EXCISION REPAIR |
|
09:10 – 09:30 |
Karl-Peter Hopfner |
STRUCTURAL ANALYSIS OF PROTEIN DNA REMODELLER |
|
09:35 – 09:55 |
Dale Wigley |
STRUCTURE AND REGULATION OF HUMAN INO80 COMPLEX |
|
10:00 – 10:30 |
Refreshments |
|
|
THURSDAY AM |
DNA Sculpting & Human Disease |
|
|
10:30 – 10:35 |
Session Introduction by Dorothy Erie |
|
|
10:35 – 10:45 |
Dong Wang * |
MOLECULAR MECHANISM OF TRANSCRIPTION TRANSCRIPTION-COUPLED REPAIR |
|
10:50 – 11:10 |
Phoebe Rice |
AN MCM-RELATED HELICASE AND OTHER REPLICATION-RELATED MACHINERY FOUND ON THE MOBILE ELEMENT BEHIND THE MRSA EPIDEMIC |
|
11:15 – 11:35 |
Sheila David |
BASE EXCISION REPAIR GLYCOSYLASES MUTYH AND NEIL1: THE GOOD, THE BAD AND THE DAMAGED |
|
11:40 – 11:50 |
Ivaylo Ivanov * |
LESION SEARCH AND BASE EXTRUSION STRATEGY OF THYMINE DNA GLYCOSYLASE |
|
11:55 – 12:15 |
John Tainer |
THE RISE OF THE MOLECULAR MACHINES WITH CANCER IMPLICATIONS |
|
12:20 – 12:30 |
Closing Comments by John Tainer & Tom Blundell |
|
FRIDAY 16TH FEBRUARY 2018 |
|
|
07:15 – 19:00 |
Group Trip: Chichen Itza (signups in advance required) |
*Selected from abstracts.
MEET OUR POSTER PRESENTERS |
|||
|
Poster # |
Name |
Organisation |
Country |
|
1 |
Noritaka Adachi |
Yokohama City University |
Japan |
|
2 |
Matthew Bochman |
Indiana University |
USA |
|
3 |
Gloria Borgstahl |
Eppley Institute for Research in Cancer |
USA |
|
4 |
Amer Bralic |
King Abduallah University of Science and Technology |
Saudi Arabia |
|
5 |
Brian Caldwell |
Ohio State Biochemistry Program |
USA |
|
6 |
Rodrigo Orlandini de Castro |
Oklahoma Medical Research Foundation |
USA |
|
7 |
Petr Cejka |
Institute for Research in Biomedicine |
Switzerland |
|
8 |
Heejin Chung |
Sungkyunkwan University |
South Korea |
|
9 |
Anders Clausen |
University of Gothenburg |
Sweden |
|
10 |
Debanu Das |
Accelero Biostructures |
USA |
|
11 |
Anthony Davis |
UT Southwestern Medical Center |
USA |
|
12 |
Ambra Dondi |
European Institute of Oncology |
Italy |
|
13 |
Pauline Douglas |
University of Calgary |
Canada |
|
14 |
Amira Fitieh |
University of Alberta |
Canada |
|
15 |
Benjamin Foster |
IFE, Helmholtz Zentrum München |
Germany |
|
16 |
Harshad Ghodke |
University of Wollongong |
Australia |
|
17 |
Morgan Hepburn |
University of Calgary |
Canada |
|
18 |
Han Ho |
University of Wollongong |
Australia |
|
19 |
JinKyung Hong |
Sungkyunkwan University School of Medicine |
South Korea |
|
20 |
Jerry Houl |
University of Texas MD Anderson Cancer Center |
USA |
|
21 |
Christopher Jackson |
Yale University |
USA |
|
22 |
Seong-Whan Jeong |
The Catholic University of Korea |
South Korea |
|
23 |
David Jeruzalmi |
City College of New York |
USA |
|
24 |
Nicholas Jette |
University of Calgary |
Canada |
|
25 |
Stanislaw Jozwiakowski |
University of Zurich |
Switzerland |
|
26 |
Jacek Kabzinski |
Medical University of Lodz |
Poland |
|
27 |
Yoko Katsuki |
Kyoto University |
Japan |
|
28 |
Yoori Kim |
The University of Texas at Austin |
USA |
|
29 |
Ina Klusmann |
Universitätsmedizin Göttingen |
Germany |
|
30 |
Rebecca Ley |
University of Sheffield |
UK |
|
31 |
Ireneusz Majsterek |
Medical University of Lodz |
Poland |
|
32 |
Lucia Francesca Massari |
European Institute of Oncology |
Italy |
|
33 |
Riccardo Miggiano |
University of Piemonte Orientale |
Italy |
|
34 |
Yeom Mina |
Sungkyunkwan University School of Medicine |
South Korea |
|
35 |
Davide Moiani |
UT MD Anderson Cancer Center |
USA |
|
36 |
Elizabeth O'Brien |
California Institute of Technology |
USA |
|
37 |
Taiana Maia de Oliveira |
AstraZeneca |
UK |
|
38 |
Richard Pomerantz |
Temple University |
USA |
|
39 |
Fahad Rashid |
KAUST |
Saudi Arabia |
|
40 |
Samuel Redstone |
University of Utah |
USA |
|
41 |
Liton Kumar Saha |
Kyoto University |
Japan |
|
42 |
Lauren Salay |
Vanderbilt University |
USA |
|
43 |
Daniel Saltzberg |
University of California San Francisco |
USA |
|
44 |
Altaf Sarker |
Lawrence Berkeley National Lab |
USA |
|
45 |
Lisa Schubert |
University of Copenhagen |
Denmark |
|
46 |
Runze Shen |
The University of Texas M.D. Anderson Cancer Center |
USA |
|
47 |
Agnieszka Anna Smolińska |
Ernst-Moritz-Arndt-University Greifswald |
Germany |
|
48 |
Lucas Struble |
UNMC Eppley Institute |
USA |
|
49 |
Michal Roman Szymanski |
University of Gdansk and Medical University of Gdansk |
Poland |
|
50 |
Roopa Thapar |
MD Anderson Cancer Research Center |
USA |
|
51 |
Marcel Tuppi |
Goethe-Universität Frankfurt |
Germany |
|
52 |
Felicity Watts |
University of Sussex |
UK |
|
53 |
Richard Wood |
The University of Texas MD Anderson Cancer Center |
USA |
|
54 |
Dongyi Xu |
Peking University |
China |
|
55 |
Yixi Xu |
Peking University |
China |
|
56 |
Zhe Yang |
The Rockefeller University |
USA |
|
57 |
Manal Zaher |
King Abdullah University of Science and Technology |
Saudi Arabia |
Supported by
Interested in sponsoring this conference?
Contact usVenue & Location
Fiesta Americana Condesa
This stylish hotel features contemporary Mexican architecture, including one of the largest and most impressive thatched-roof "palapas" in the entire country. There are cultural activities, arts and crafts and sports programs to keep you constantly entertained, plus time to relax and enjoy the Mayan culture, soak up the Caribbean sunshine and revel in the international ambiance that settles in after the sun goes down.
Throughout your stay delegates will enjoy a full meal plan, inclusive of beverages. Take your pick from the aromatic Asian delights at Kaumbu, traditional fare at El Mexicano, delicious international cuisine at Kalmia Buffet or perhaps sample the sumptuous Italian dishes at Rosato. There are also several other dining opportunities such as the Cevichería, Pizzeria, Sushi Corner, pool and lounge bar areas. The Gala Night with either a Mayan or Caribbean theme takes place on the third evening of the conference with a mouth-watering feast of local cuisine, an open bar and amazing local entertainment. We welcome all delegates and their accompanying persons to the Gala Night – a truly fun filled night not to be missed!
Hotel Facilities
- Lagoon Pool
- Extensive Health Club and Spa
- Complimentary Wi-Fi in guest rooms and throughout hotel and conference areas
The Fiesta Americana Condesa, Cancun is the 2015 Winner of the Trip Advisor Certificate of Excellence and also received the Travellers Choice Award in 2014.
The remains of ancient Mayan cities are scattered throughout the Yucatan Peninsula and no trip to Cancun would be complete without a visit to these majestic temples and pyramids set amongst lush tropical vegetation. We will be working very closely with a reputable tour company who will be organising trips to Tulum, Chichen Itza, Coba and Xcaret to name a few, some of which may require a full day. Full day excursions will be arranged pre or post conference and may be on an individual or group booking so we would recommend booking extra nights to extend your stay as early as possible to avoid disappointment should these excursions be of interest to you and/or your party. For further information please contact us.
General Information
Venue Rating
★ ★ ★ ★ ★
Currency
US Dollar (USD)
Address
Fiesta Americana Condesa, Blvd.Kukulkán km 16.5 Zona Hotelera, Cancun, 77500 Mexico
Nearest Airport
Cancun International Airport
Location
Cancun is a delightful combination of natural beauty, islands, ecological reserves and white sandy beaches. However, besides sun, sand and sea, this destination also offers an infinite variety of underwater activities to choose from: the diving, snorkelling and fishing here are outstanding and you will find an undersea world packed with tropical fish that live on the second largest barrier reef in the world. Sports enthusiasts might choose one of the many eco-tourism activities, such as cycling or hiking through the tropical forest or kayaking through mangroves, or something a little more adventuresome, like zip lining through the treetops.
Apart from the more well-known ancient Mayan archealogical sites such as Tulum, Cobá and Chichén Itzá you may like to visit the Aktun Chen caverns, voted one of the Top 10 underwater walks by National Geographic described as a truly magical experience.
Gallery
View on FlickrConference History
If you are interested in this meeting but not yet ready to register, you can sign up for updates here and our team will keep you updated regarding deadline reminders and grant opportunities relating to this meeting only.
If you're interested in sponsoring this conference please contact us.
Conference Manager

Laura Trundle


Need some help? Chat to the Fusion team today
As a family run business, our dedication runs deep. We’re committed to each other and, even more so, to every attendee’s experience, delivering a level of care and passion that’s truly unmatched.


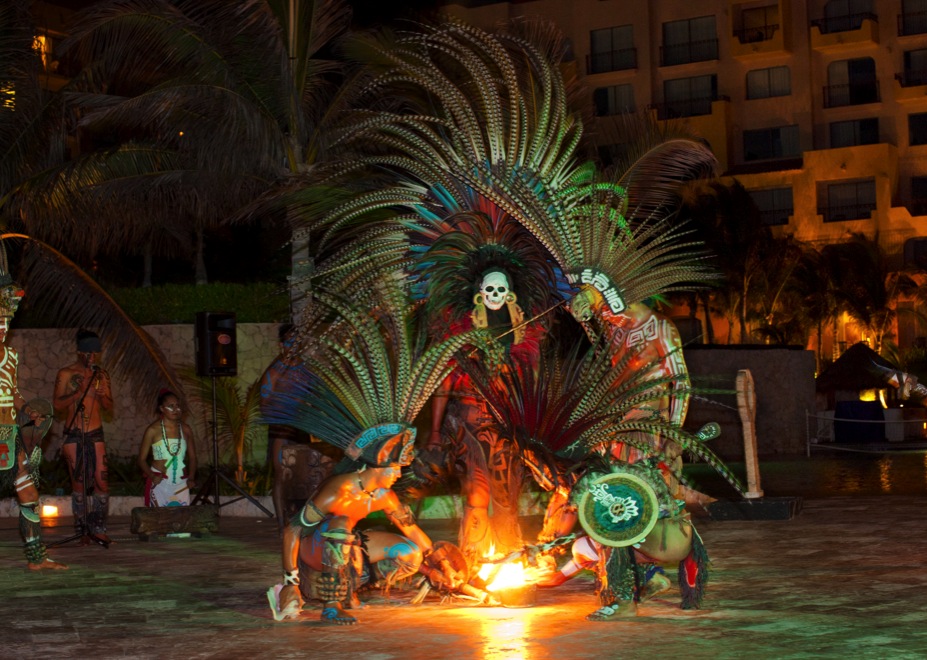


















.png)